Mnova 是西班牙Mestrelab Research公司開發的 核磁數據處理軟體,集NMR 和LC/MS 數據處理分析、預測、發表、驗證及數據的儲存、檢索以及管理等功能於一身,具有功能強大健全、支持無縫共享數據,可以為用戶提供準確的數據處理。具操作簡便人性化、處理結果準確美觀等優勢。
更新介紹
MNOVA 14 – HIGHLIGHTS
1- Mnova ElViS for Electronic and Vibrational Spectroscopies
2- Mnova BioHOS for the determination of a biologic drug’s higher order structure
3- Mnova Gears for automating your analytical workflows with already 11 Gears bricks
4- Mnova StereoFitter for 3D conformational and configurational analysis
5- A new Ensemble NMR Prediction that uses several prediction algorithms
6- 2D NMR Resolution Booster: A new algorithm for the resolution enhancement of 2D NMR spectra
7- qNMR and Concentration determination: Improved usability, reporting, and many new features
8- Mnova Screen v1.3.
9- Data Analysis panel – stack chromatograms from MS or UV detectors
10- Compliance tools such as Digital signatures & Audit trail
11- Mnova NMR – Advised Processing tool: The most sensible processing options for most 1D and 2D NMR spectra
12- Mnova NMR – NMReData: Export NMR information following the new NMReData standard format.
13- Mnova NMR – NMR VOI compression: A new algorithm for the efficient VOI compression of NMR spectra
14- Several new features for Mnova MS such as improved default settings for uploading MS data or labels can be added to mass spectrum peaks and datasets
Expand Mnova Capabilities
Plug-ins, basic and advanced
| BASIC |  |
An outstanding suite to visualize, process, analyze and report your data. |
 |
Process, analyze and report your Mass Spec and/or Chromatographic data. | |
 |
Carry out the analysis of various optical spectroscopy data. | |
| ADVANCED |  |
Accurate prediction of 1H and 13C NMR spectra from a chemical structure and others |
 |
Automatic confirmation of structure identity based on NMR and/or LC/GC/MS data. | |
 |
Assisted NMR quantitation! Concentration or purity determinations. | |
 |
Store, share and search your chemical and analytical data efficiently. | |
 |
Extraction of spectroscopic and chemical kinetic concentration from arrayed NMR datasets. | |
 |
A state-of-the-art automatic analysis tool for ligand screening NMR data. | |
 |
Quantitation of mixture components by NMR based on chemical shift ranges. | |
 |
Validated and quantitative structure-property prediction integrated in Mnova! | |
 |
From NMR data to structure elucidation in a simple and robust workflow. | |
 |
Chemical shift perturbation analysis for fragment-based drug discovery. | |
 |
IUPAC names can be generated for structures from Mnova 12. | |
 |
Designed to assist with the analysis of the NMR spectra of biotherapeutics. | |
 |
Build automation workflows for your analytical data & more | |
 |
3D conformational and configurational analysiss from NMR spectra | |
 |
Efficient batch processing tools for lead discovery using protein-observed 2D NMR |
特色
PLUG-INS
Mnova has 3 basic plugins covering several techniques: Mnova NMR, Mnova MS and Mnova ElViS . In addition, Mnova can run a number of additional advanced modules such as mixtures analysis, reaction monitoring, quantitation, chemical shift prediction, screening, verification as well as physico-chemical properties prediction.
ONE SHARED INTERFACE
Mnova combines all your analytical data within the same software interface. This is a time saver and a very efficient way to process, analyze and report your data.
ONE CORE APPLICATION
Mnova enables you to increase your productivity by combining your analytical data in one piece of software. Advanced algorithmia runs in the background to provide quality results.
WORKFLOW OPTIMIZATION
Excellent ability to manually interact with your data as well as to prepare an automated workflow to save your precious time.
SCRIPTING ENGINE
Perform any action available in the Mnova graphical user interface but in full automation. Our developers have created big projects using our scripting engine and we can help your business if you have specific needs.
MINIMAL LEARNING CURVE
Bringing all your analytical data into one single interface (powerpoint look) makes Mnova a piece of software with an extremely quick rate of learning.
|
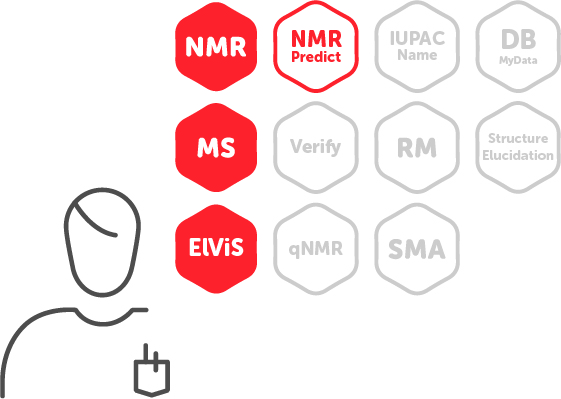
Mnova Suite |
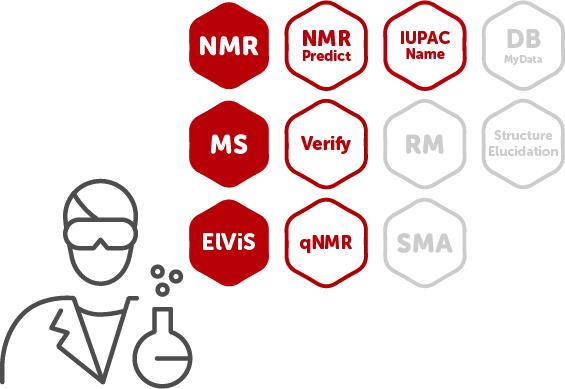
Mnova Suite Chemist |
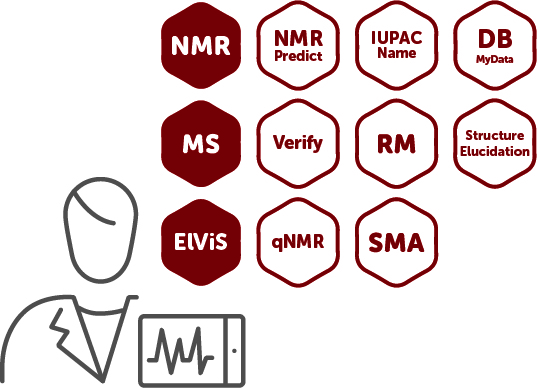
Mnova Suite Expert |
 |
 |
 |
This is the ideal entry package for users who want to process and visualize their data in a single software tool, whether this is NMR or MS. With this package you will be able to open, view, process, analyze, report and predict NMR and MS data, from multiple vendors. |
This combo is designed for synthetic chemists. It has all the processing and visualization power of Mnova Suite as well as the tools to automatically confirm your structure, and obtain the purity or concentration for your compounds of interest. |
The most complete package developed by Mestrelab in the Mnova environment, designed for experts analytical chemists. You can do everything, plus take advantage of databasing, assisted de novo structure elucidation, follow reactions, extract kinetics and much more! |
| NMR + MS + ElViS + NMRPredict | NMR + MS + ElViS + NMRPredict + Verify + qNMR + IUPAC Name |
NMR + MS + ElViS + NMRPredict + Verify + qNMR + DB MyData + RM + SMA + IUPAC Name + Structure Elucidation |
Windows System Requirements
The minimum recommended configuration for Mnova installation is at least 1 GHz or faster processor, 1 GB RAM, a VGA color monitor with 800 x 600 pixels resolution, a compatible mouse and Windows 7 as operating system. However, ideal system requirements for optimum operation of the software are Pentium 1,6 GHz or higher CPU with 2 GB or more of RAM memory and Windows W10 or 11.
Mnova will not install under Windows XP or lower. The default installation requires about 1,1 GB of disk space. If you are going to install (or uninstall) the program on a Windows system, be sure you have administrator privileges, because the installation procedure is going to install several files in your system folders.
If you don't have such privileges, you won't be able to install or uninstall the program correctly.
Mac OS X System Requirements
Mnova requires at least Mac OS X 10.13
Linux System Requirements
Mnova requires at least Pentium 1 GHz, 1 GB RAM, Video Adapter Super VGA (800 x 600) with X Library (Xlib). Please, make sure to choose the correct setup file for your Operating System (Debian, OpenSUSE, Fedora, Ubuntu, Red Hat Enterprise...)


